Summary
Genetic engineering can be very useful not only in agriculture, medicine and other fields. The example of the American chestnut shows that it can also be very helpful in ecology. Genetic engineering is certainly not a “silver bullet” to solve all problems. TASF and ESF pursue all possible avenues simultaneously in order to find the best solution.
Note the addendum (Jan. 16th, 2024) at the end of the article!
The American chestnut (Castanea dentata) was an impressive tree. With a height of up to 45m and a trunk diameter of up to 3m, it dominated the forests of the eastern USA from Maine to Mississippi and was of great ecological importance: the crown with a diameter of 30m provided shade, protected against desiccation and fixed CO2. The fruits served as food for wild animals such as turkey, bear, deer and others. Furthermore, the fast-growing tree supplied valuable hardwood, used for both furniture and sustainable building material.
Why do we speak in the past tense?
In 1904, “bark canker,” a fungal disease (Castanea dentata), was introduced with the Chinese ornamental chestnut. Within a few decades, the fungus destroyed almost the entire population of 3.5 billion chestnut trees.

Infected stem of American chestnut.
The fungus blocks the water supply of the tree and essentially “strangles it to death”.
What could be done?
“The American Chestnut Foundation” (TACF) is working with scientists at the State University of New York College of Environmental Science and Forestry (ESF) to try and save the chestnut in several ways:
1. it is possible that among the few surviving trees there are a few that have developed resistance against the fungus. It is a very difficult, lengthy procedure to identify them – and with very uncertain success. Many mature American chestnuts were cut down to be used for lumber before the blight could kill them. This premature harvest killed off much of the genetic diversity of the American chestnut population that could have contained the genes for natural resistance. Most “resistant” trees that are found have generally just avoided infection from the blight due to their geographical location. These “surviving” trees often succumb to the blight once the fungus reaches them. Older trees are more likely to be infected since the fungus invades via cracks and injuries in the bark.
2. a second approach is a cross between the American chestnut and the resistant Chinese subspecies. The hybrid should then get back as much of the genetic material of the original subspecies as possible. This can be done by many backcrosses. Because there is more than one gene that gives the Chinese chestnut blight resistance, it is laborious and time consuming to identify plants with the highest content of the original genome without losing the resistance.
3. a third approach can at least maintain the genetic diversity of the population but has other limitations. The fungus does not attack the roots of the tree. The stumps resprout and grow into small trees. However, these become infected again after a few years and die. These “dwarf trees” cannot restore the ecosystem.
4. a fourth approach seeks a genetic engineering solution. The American Chestnut Foundation has been working with SUNY ESF to develop a blight-tolerant transgenic chestnut, which we describe in a little more detail:
Darling 58
Resistance or tolerance to pathogenic fungi is known from a variety of plants. In some plants, the defense is based on oxalate oxidase (OxO), an enzyme that neutralizes oxalate. Oxalic acid is produced by the fungus and cause the cankers that eventually lead to the death of the tree. OxO allows the trees to break down oxalic acid produced by the fungus.
The enzyme catalyzes the following reaction:
Fig. 2
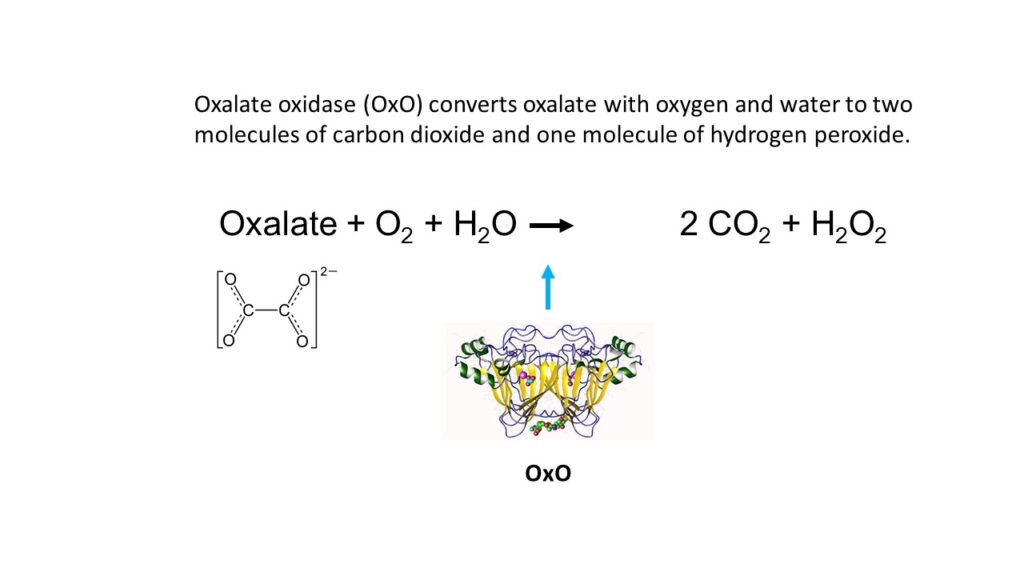
OxO occurs naturally in wheat, barley and other crops. SUNY ESF scientists selected the gene for OxO from wheat and genetically engineered it into the chestnut. This is how “Darling 58” was created. (Named, by the way, after Herb Darling, founder of the New York chapter of TACF).
How is it done?
First, cells from the plant are propagated in cell culture in the lab.These cells are then treated with Agrobacterium tumefaciens. The bacterium is a natural gene delivery vehicle for plants and can introduce DNA into the plant genome. Actually, Agrobacterium transforms plants with its own genes and creates a tumor.
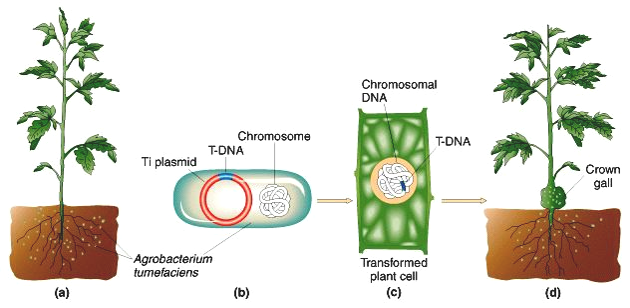
Fig. 3
In this natural infection by Agrobacterium tumefaciens, tumor genes from the Ti plasmid are introduced into the plant and cause a crown gall tumor. Replacing the tumor genes by the OxO gene will introcuce this into the plant genome and not cause a tumor.
Note: this is called the “old” or “conventional” gene technology. The new gene technology (NGT) by CRISPR-Cas cannot yet be routinely used to introduce new genes into plants.
Back to the chestnut
Unlike animals, you can grow whole plants from single cells in cell culture.

Fig. 4
After transformation by Agrobacterium, a callus, a mass of undifferentiated cells is formed. This can be induced to form leaves and roots and finally a complete plant can be generated.
One leaflet of these plantlets is sufficient to check by molecular biology that the desired gene, in this case OxO, has been incorporated and that no other changes have taken place. Then young saplings are infected with Castanea dentata and observed how they grow. Finally, the variety Darling58 was selected, since it showed enhanced tolerance of the blight fungus.
And now, everything is fine?
With Darling 58, there was now a chestnut that contained only one additional gene (the OxO gene) and was otherwise genetically identical to the American chestnut. This is different from a cross between the American and Chinese chestnut, where two related but still very different genomes of about 40.000 genes each are thrown together.
In contrast to crosses, genetically modified plants have to be thoroughly tested according to safety regulations. for many possible side effects. Some of these tests are shown in the table.
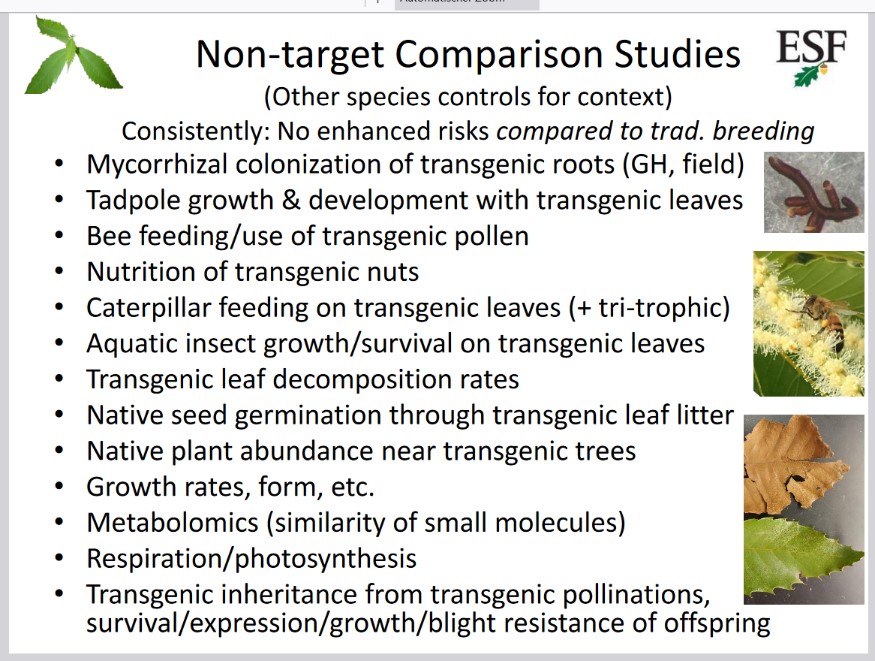
Fig. 5
List of some tests carried out to examine safety of the transgenic chestnut.
All of these tests have shown no significant difference between Darling 58 and non-transgenic American chestnuts. How complicated and lengthy such an approval process is summarized here. This process has not been completed yet. The first planting of the blight-tolerant chestnut outside of permitted sites, originally scheduled for 2019, had to be postponed. At least in part, this can be explained by the strong opposition from GM opponents.
Opposition
When science suggests gene technology solutions for a problem, opposition is not far (e.g. here and here).
The arguments are always the same: the risks of genetic engineering cannot be predicted, there is no scientific evaluation of long-term effects and the consequences on the ecosystem are unknown. This is all correct but …
The hybridisation program (crossing of Asian and American chestnuts) which is favored by the anti-GMO opposition is similarly problematic: these subspecies would normally not hybridize since they live in different ecosystems. The risks of hybridization are unknown, there are no data on long-term effects and the consequences on the ecosystem cannot be predicted. And there is one essential difference: in the hybridization program, two times 40.000 genes are randomly mixed while just one single gene is added in the GMO tree. What happens by crossing subspecies is explained in our blog post „Mutants“
Interestingly, the Sierra Club, which is essentially opposed to all genetic modifications, has taken sides with the GMO trees. The club sees a good chance of restoring the original ecosystem and was convinced mainly by the fact that the tolerant chestnut will not be patented.
Addendum, Jan. 16th, 2024
Reports in the Washington Post (long text) and at the “Genetic Literacy Project” (short text) report on a huge setback in the “American Chestnut Project”.
Apparently seeds had been mixed up or mislabelled before planting. Instead of Darling 58, a mixture of Darling 54 and Darling 58 was planted! The young Darling 54 trees are less robust, have slower growth, die more often and also seem to have a lower tolerance to the fungus.
What could be the reason for this?
We can only make assumptions here, but these are based on research experience.
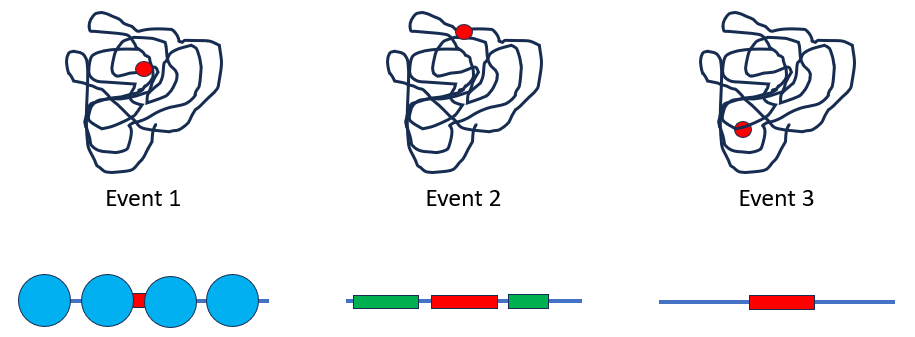
Darling 58 and Darling 54 are different “events”, i.e. the results of independent transformations (genetic engineering experiments). Both contain the oxaloxidase gene as a “transgene”, but it can be inserted at different locations in the genome. This is a disadvantage of genetic engineering with Agrobacterium: the insertion into the target genome occurs randomly, the location cannot be predicted.
Therefore, many different events are analysed and the best ones are selected.
Potential differences between Darling 58 and Darling 54
If the oxaloxidase gene is inserted into an epigenetically repressed area, it may be expressed less strongly or not at all and thus lead to less or no tolerance to the fungus (Event 1). If the transgene happens to be inserted in the immediate vicinity of genes that contribute to growth and robustness of the tree, it can disrupt or even prevent their expression (event 2). This could explain why the trees are smaller and weaker or die more often. In the best case, the transgene is inserted into a neutral region of the genome (event 3): it is properly expressed and does not interfere. This is not unlikely because, surprisingly, the vast majority of a genome does not code for genes.
Another hypothesis is based on the fact that Darling 58 carries the oxaloxigenase gene on only one of the parental chromosomes, whereas Darling 54 carries it on both. The resulting high expression could delay growth.
For the more advanced: unusually high gene expression can sometimes lead to the opposite effect in plants. This was recognised very early on in plant research and referred to as “transgene silencing”. The mechanism has now been elucidated and is called “RNA interference”. An article on this is in preparation.
So there was probably a good reason why Darling 58 and not Darling 54 was originally selected for cultivation. The apparent confusion between the two events does not result in any risk for the environment or human health . However, it not only costs a lot of time and money, it is embarrassing and also leads to a loss of confidence among the funding supporters of the project – and it is of course grist to the opposition’s mill.
We can only hope that this disastrous mix-up will be cleared up quickly and completely and that the project can be completed successfully after all, albeit with a delay.
Acknowledgement
Thanks to Adriana del Grosso and Andy Newhouse from The American Chestnut Research and Restoration Project at SUNY College of Environmental Science and Forestry (ESF), for providing figures and valuable comments on the text.
Author: Wolfgang Nellen, BioWissKomm
Cover image: BioWissKomm, created by Midjourney
Translation by DeepL with editing by the author
OxO-figure: BioWissKomm
all other figures kindly provided by ESF

